GinDB-AI platform, a comprehensive resource that offers detailed information regarding ginsenosides, their spectrometric and chromatographic parameters, and their biological functions. This platform is equipped with artificial intelligence (AI)-based predictive tools designed for the analysis of multidimensional molecular features related to ginsenosides and their biological activities. We systematically compiled data on ginsenosides and their associated information from the literature spanning 1963 to 2024, alongside reputable databases such as PubChem and ChEMBL, to construct the ginsenoside database (GinDB). We utilized ChemDraw and RDKit to generate the missing molecular strings (canonical SMILES) and structural representations, thereby enriching the database with essential information about ginsenosides. Based on this curated database, we developed AI-based predictive tools capable of predicting physicochemical properties and biological activities of ginsenosides. These tools comprise two regression models and a classification model, developed utilizing our proposed deep learning (DL) architectures, incorporating conventional features and natural language processing (NLP) embeddings.
Reference: GinDB-AI: An integrated ginsenoside database and AI-driven platform for multidimensional information and biological activity prediction
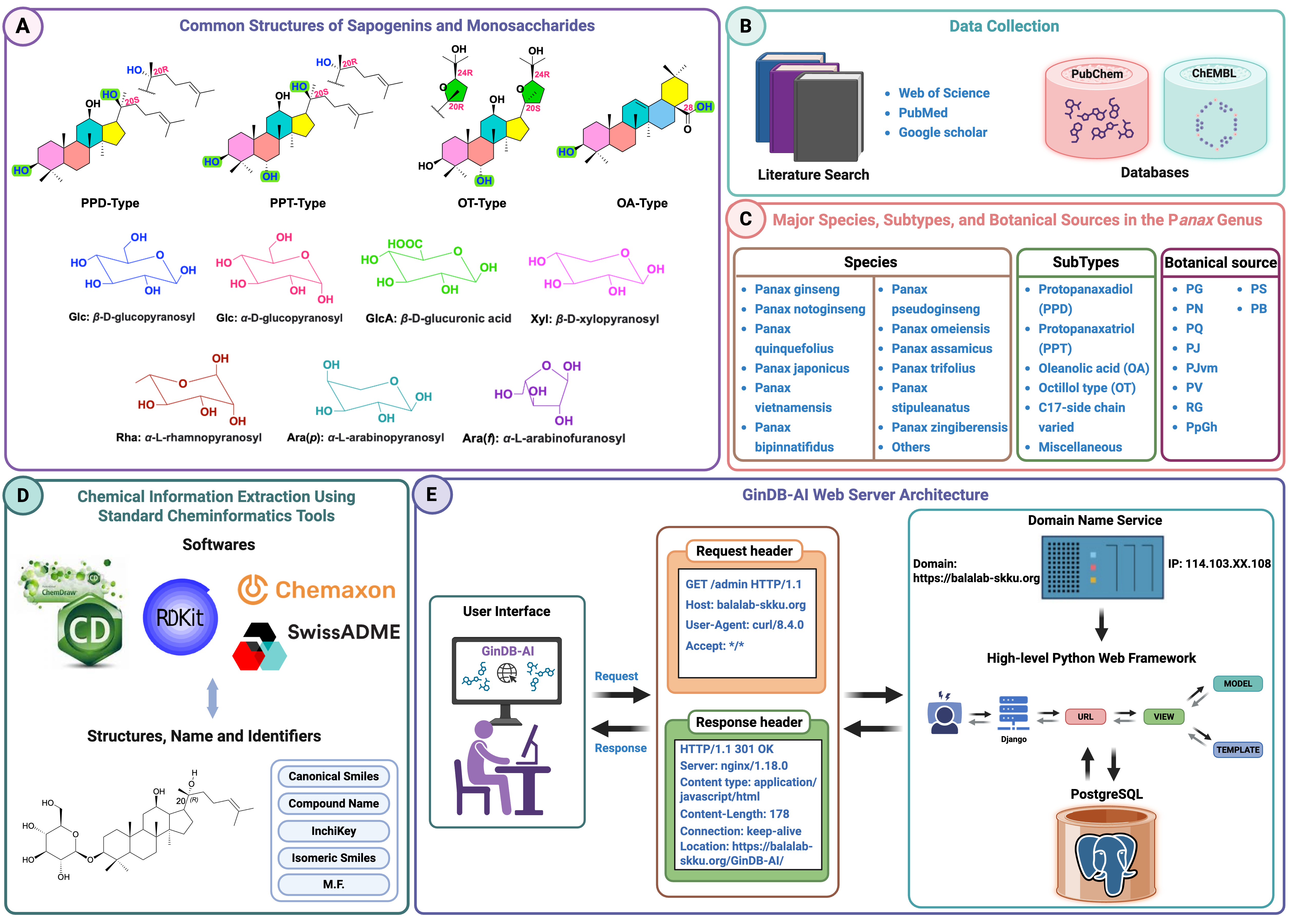
Aim and Objective: The primary aim of GinDB-AI is to assist the scientific community engaged in ginsenoside research, functional food development, and natural product-based drug discovery. This platform compiles and centralizes scattered information on ginsenosides, including their chemical structures, physicochemical properties, chromatographic and spectrometric data, and biological activities. Our objective is to provide both bioinformatics and experimental researchers with a reliable, AI-driven resource for retrieving, analyzing, and predicting multidimensional information and functional potential of ginsenosides.
Search Options: The database offers i) Basic Search which allows the user to search in any field or against multiple fields. By default, it searches against major fields. It also allows the user to DISPLAY desired fields of the database. ii) Advanced Search allows user to construct queries in order to search the database using multiple queries joined by logical operators like AND/OR/NOT. This search module allows user to perform search on any or all fields of the dataset. It also allows to display all field or user selected fields.
Browse: Modules under this section allow users to retrieve data in a structured manner. These modules enable browsing of ginsenoside compounds and their related information based on various properties, categories, and functional classifications.
Tools: Modules under this section integrate various web-based tools for predicting multidimensional information of ginsenosides and classifying their biological functions.
Mobile Compatible Website: GinDB-AI is built using bootstrap framework so the website is responsive and is compatible for desktop, tablet, and smartphone.